One of the challenges in toxicology is the ability to extrapolate the results from different stages of risk analysis, from experimental systems to human populations. Animal models in particular, although widely used, often have differences, particularly in terms of substance clearance or enzymatic activity. For these practical reasons, but also for ethical, political, and economic considerations, significant efforts are required from laboratories and industries to Replace these models with other alternatives, Reduce their use, and Refine experimental strategies to minimize animal stress and pain (the principle of “3R”). It is in this context that chemical risk prevention and management organizations are turning to computational toxicology and more specifically to predictive toxicology, which aims to minimize the number of in vivo experiments on animals by replacing them with in vitro or in silico methods to evaluate risks to humans, as accurately if not more so.
The use of predictive toxicology is particularly recommended in the context of the REACH legislation. Predictive toxicology involves determining the toxicological “signature” of a compound using different models (animals, in vitro, in silico) and extrapolating it to predict its effect on humans and the environment. This signature can be established at different levels (physiological, molecular, genomic…), on an individual or its offspring, after exposure to one or more factors (biological, physical, or chemical). The global techniques known as “omics”, which include genomics (study of the genome), transcriptomics (study of gene expression), proteomics (study of proteins), and metabolomics (study of metabolites), are increasingly used qualitatively and quantitatively to characterize the different states of biological systems.
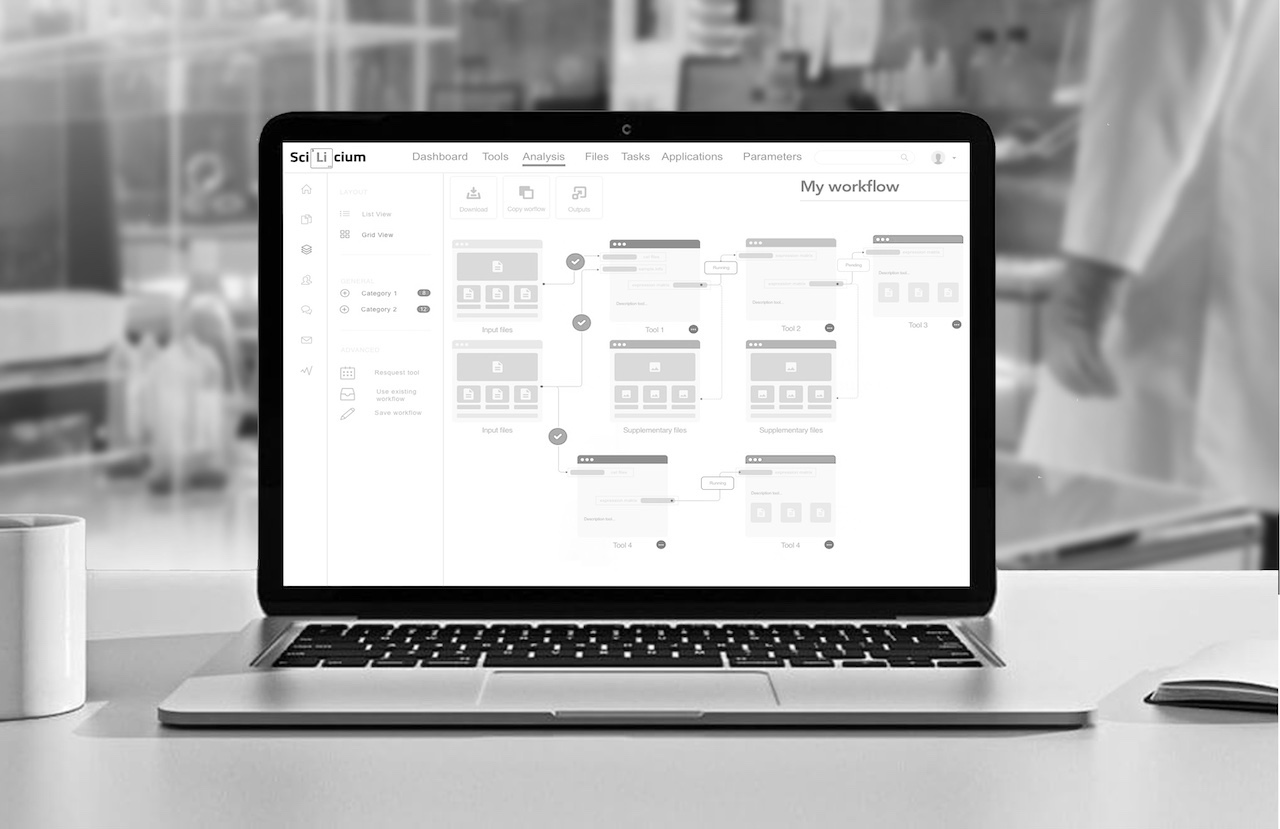
The application of these recent technologies to toxicological risk assessment has given rise to toxicogenomics, which aims to study the activity of the genome (genes, proteins) after exposure to a xenobiotic, in order to develop global, rapid, and relatively low-cost decision-making procedures for risk assessment. Today, toxicogenomics is mainly based on high-throughput sequencing techniques such as RNA-seq, allowing a deeper analysis of the genomic impact of a compound. However, toxicogenomic data analysis requires expertise in both bioinformatics and toxicology. In its various projects, SciLicium has developed an automated transcriptomic data pipeline (using BRB-seq technology). This pipeline is based on a precise sequence of analysis and interpretation tools orchestrated by a SnakeMake workflow manager.



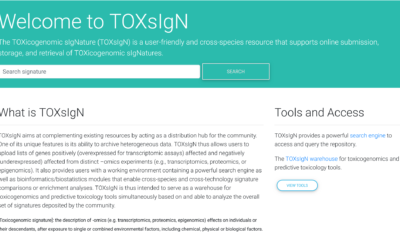
We are happy to announce that, SciLicium and the GenOuest bioinformatics platform have been granted by the Brittany region to transfer the competencies of Genouest in cloud computing to help Scilicium to develop its toxicogenomic signature analysis platform (TOXInCloud project).
This platform will attempt to be used as a predictive toxicogenomics tool in various ways. For example, it can help identify potential biomarkers for toxic exposure, which can be used to diagnose and monitor the effects of toxic agents on an individual. The platform could also help identify mechanisms of toxicity, which can be used to develop better treatments for toxic exposures.
Overall, this toxicogenomic signatures analysis platform in toxicology provides a valuable tool to assess the effects of toxic agents on an organism at the molecular level to improve the safety and efficacy of chemicals, drugs, and other agents, and ultimately protect human health.



0 Comments