Let’s talk about RNA
RNA sequencing is a technique used to analyze entire genomes by examining the expression of their genes. Such genome expression analysis becomes a standard tool for genomic studies.
Nonetheless, RNA sequencing is still expensive and time-consuming. It requires an expensive preparation of a complete genomic library (the pool of DNA generated by the RNA of cells). And finally, the generated data themselves are also difficult to analyze. All of this makes RNA sequencing difficult to perform, making its adoption less widespread than it could be.
These last years, new approaches have emerged, spurred by the revolution in single-cell transcriptomics, which uses what is known as “sample barcode” or “multiplexing”. Individual barcode sequences are added to DNA fragments during library preparation allowing the fragment identification before the final data analysis. This approach requires only one library that contains multiple samples or separate cells.
Barcoding reduces both cost and time, which could extend to bulk RNA sequencing of large sets of samples. But there are still challenges in adapting and validating protocols for reliable and inexpensive profiling of bulk RNA samples.
Bulk RNA Barcoding and sequencing” (BRB-seq)
In 2019, scientists from Bart Deplancke’s laboratory at the EPFL Institute of Bioengineering developed a new approach called “Bulk RNA Barcoding and sequencing” (BRB-seq) which is 25 times cheaper than classic commercial RNA sequencing technology (Illumina’s TruSeq).
Among its many advantages, BRB-seq is fast and preserves strand specificity which is a challenge in the field, related to transcribing DNA in the right direction. Thus, BRB-seq offers an inexpensive approach to perform transcriptomics on hundreds of RNA samples, which can increase the number of biological replicats (and therefore the experimental precision) in a single run.
In terms of performance, the scientists found that:
- BRB-seq can detect the same number of genes as the “gold standard” in the field, namely TruSeq Stranded mRNA, at the same sequencing depth.
- The technique produces reliable data even with low-quality samples.
- It generates genome-wide transcriptomic data with a cost similar to the profiling of four genes using RT-qPCR (low-throughput method for measuring gene expression).
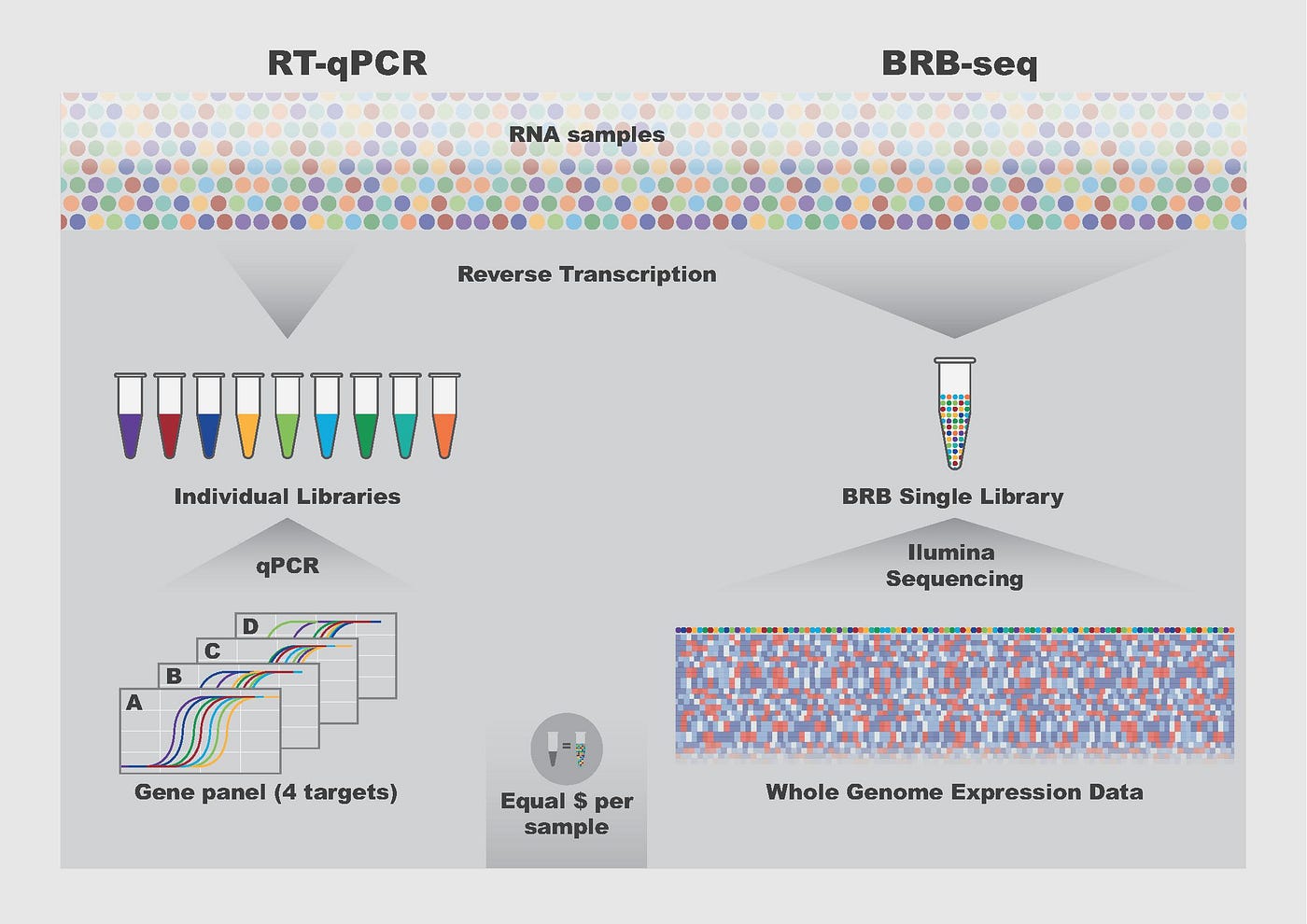
In one test, BRB-seq could generate genomic libraries ready to sequence up to 192 samples per day, requiring only two hours of hands-on time. This technique is combined with a user-friendly pipeline for the preprocessing and analysis of sequencing data, allowing the acquisition of results in a single day.
According Bart Deplancke:
Since its launch, dozens of laboratories and companies have already contacted us to help them implement the BRB-seq approach. Due to the low cost of BRB-seq, these researchers realized that they could now analyze many more samples with the same budget, greatly increasing the scope and reproducibility of their experiments. We, therefore, expect the BRB- seq or a comparable approach will in the long run become the norm in any molecular biology laboratory and replace RT-qPCR as the first option for gene expression profiling.




0 Comments